How to submit jobs to clusters
Anton
- mv step5_md-1.gro md.gro
- run prep1-3.sh, you will get out.dms
- run step1-3.sh, job will be submitted
Notes:
- Anton uses kcal/mol/A
- 1.2 was used to do position restraints, ~500 in Gromacs
- ff99SB-starIDLN is self-contained
- ff14SB only has protein, used OL15 for DNA, OL3 for RNA, TIP3P for water, ions.amber1234lm_anton.tip3p for ions
To continue a simulation upto timestep X, simply
anton2 submit jobid --cfg anton.tune.last_time=500000
Most updated force-fields
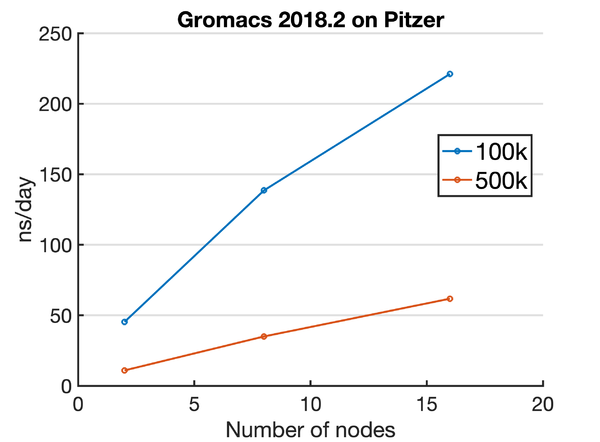
Pitzer
We participated in the Pitzer Early Program, which will become open to the public from Dec 4 2018.
Here is a page on migrating your job from Owens to Pitzer.
Introduction to Pitzer queues and batch limits
Here is a standard Pitzer Gromacs job script:
#PBS -N Ups1-NN
#PBS -l nodes=10:ppn=40
#PBS -l walltime=24:00:00
#PBS -S /bin/bash
#PBS -j oe
#PBS -A PAS1326
trap "cd $PBS_O_WORKDIR; rsync -avh $TMPDIR/ .; exit" TERM
date
module load gromacs
# PBS_O_WORKDIR refers to the directory from which the job was submitted.
cd $PBS_O_WORKDIR
prefix=step5_md
gmx convert-tpr -s ${prefix}-MM.tpr -o ${prefix}-NN.tpr -extend 200000
pbsdcp -p ${prefix}-MM.cpt ${prefix}-NN.tpr $TMPDIR
# Use TMPDIR for best performance.
cd $TMPDIR
mpiexec gmx_mpi mdrun -v -noappend -ntomp 1 -cpi ${prefix}-MM.cpt -deffnm ${prefix}-NN
pbsdcp -p * $PBS_O_WORKDIR/
Things to note:
- The trap command is for copying out files when terminating
- -noappend is required for Gromacs/2018.2 which is default on Pitzer
- -ntomp 1 is A MUST on both Owens and Pitzer, might not be stated on the software page but confirmed by the OSC staff
Submit dependent jobs using this