How to submit jobs to clusters
Contents
Schedule
Date in the bracket indicates the end date.
| Machine | GPU0 | GPU1 | GPU2 | GPU3 |
|---|---|---|---|---|
| uc62 | Shen[02/28] | |||
| uc63 | Zoe | |||
| uc64 | Rui 2/24 | Sijie 2/24 | ||
| uc65 | Xiaotian | |||
| uc66 | ||||
Exxact
Make sure you first create a directory (using your name) under /data and run everything in there. DO NOT create things under /home/exx. For example:
mkdir /data/kevin cd /data/kevin
Amber18 was pre-installed under /usr/local/amber18/. To run CUDA version of Amber, simply
export CUDA_VISIBLE_DEVICES=0 pmemd.cuda -O -i 06_Prod.in -o 06_Prod.out -p ionized.parm7 -c 05_Pull.rst -r 06_Prod.rst -x 06_Prod.nc
If you would like to simultaneously utilise multiple GPUs, even simpler
export CUDA_VISIBLE_DEVICES=0 pmemd.cuda -O -i 06_Prod.in -o 06_Prod.out -p ionized.parm7 -c 05_Pull.rst -r 06_Prod.rst -x 06_Prod.nc export CUDA_VISIBLE_DEVICES=1 pmemd.cuda -O -i 06_Prod.in -o 06_Prod.out -p ionized.parm7 -c 05_Pull.rst -r 06_Prod.rst -x 06_Prod.nc
To check status of the GPUs
$ nvidia-smi
Currently we have to rely on manual scheduling of running jobs among group members. If you need a node or two, feel free to ask me! -Kev
Anton
- mv step5_md-1.gro md.gro
- run prep1-3.sh, you will get out.dms
- run step1-3.sh, job will be submitted
Notes:
- Anton uses kcal/mol/A
- 1.2 was used to do position restraints, ~500 in Gromacs
- ff99SB-starIDLN is self-contained
- ff14SB only has protein, used OL15 for DNA, OL3 for RNA, TIP3P for water, ions.amber1234lm_anton.tip3p for ions
To continue a simulation upto timestep X, simply
anton2 submit jobid --cfg anton.tune.last_time=500000
Most updated force-fields
$ garden with -m viparr/4.7.12c7/bin -m viparr-ffpublic/1.0.3c7/data — viparr —help
usage: viparr input.dms output.dms [ options ]
VIPARR version 4.7.12
VIPARR_FFPATH: /gdn/centos7/0001/x3/prefixes/viparr-ffpublic/1.0.3c7__e02630aad12e/ff
All available forcefields ————————————— aa.amber.ff03r1 — Amber ff03r1 protein forcefield aa.amber.ff14SB — Amber ff14SB protein parameters aa.amber.ff15fb — Amber ff15fb protein parameters aa.amber.ff15ipq — Amber ff15ipq protein parameters aa.amber.ff94 — Amber ff94 protein forcefield aa.amber.ff96 — Amber ff96 protein forcefield aa.amber.ff99 — Amber ff99 protein forcefield aa.amber.ff99SB — Amber ff99SB protein forcefield aa.amber.ff99SB-ILDN — Amber ff99SB-ILDN protein forcefield aa.amber.ff99SB-disp — Amber ff99SB-ILDN protein forcefield with additional vdw and torsion modifications. For use with water.tip4pd-1.6 model aa.amber.ff99SBnmr — Amber ff99SBnmr protein forcefield aa.amber.ff99SBstar-ILDN — Amber ff99SB*-ILDN protein forcefield aa.amber.ffncaa — Amber protein forcefield for 147 non-canonical amino acids aa.amber.ffptm — Amber protein parameters for 32 common post-translational modifications aa.amber.phosaa10 — Amber phosphorylated amino acids aa.charmm.c22 — Charmm22 protein forcefield aa.charmm.c22der — Charmm22 protein forcefield with modified DER side chain charges aa.charmm.c22h — Charmm22 forcefield with pseudopolarization term aa.charmm.c22nocmap — Original charmm22 protein forcefield without added cmap terms aa.charmm.c22star — Charmm22 protein forcefield with backbone and sidechain torsion and DER charge modifications aa.charmm.c36 — Charmm36 protein forcefield aa.charmm.c36m — Charmm36 protein forcefield (c36 with additional cmap and NBFIX modification) carb.charmm.c36 — Charmm36 carbohydrate forcefield ethers.charmm.c35 — Charmm35 ether forcefield ions.amber1234lm_anton.spce — Amber mon- di- tri- and tetra- valent ion parameters for use with spce. Stable on Anton ions.amber1234lm_anton.tip3p — Amber mon- di- tri- and tetra- valent ion parameters for use with tip3p. Stable on Anton ions.amber1234lm_anton.tip4pew — Amber mon- di- tri- and tetra- valent ion parameters for use with tip4pew. Stable on Anton ions.amber1ff99.tip3p — Original Amber ion parameters contained in the ff99 forcefield ions.amber1jc.spce — Amber monovalent ion parameters for use with spce ions.amber1jc.tip3p — Amber monovalent ion parameters for use with tip3p ions.amber1jc.tip4pew — Amber monovalent ion parameters for use with tip4pew ions.amber1lm_iod.all — Amber monovalent ion parameters ions.charmm22 — Original ion parameters used with charmm forcefields. ions.charmm36 — Ion parameters used with charmm36 forcefields. Includes NBFIX terms lipid.amber.lipid14 — Amber Lipid14 parameters lipid.amber.lipid17 — Amber Lipid17 parameters lipid.charmm.c36 — Charmm36 lipid forcefield misc.charmm.all36 — collecton of most charmm36 templates and parameters in one spot na.amber.OL15 — Amber DNA parameters, OL15 variant na.amber.OL3 — Zgarbova et al. (2011) JCTC 7: 2886 na.amber.ROC — Amber RNA parameters, ROC variant na.amber.YIL — Amber RNA parameters, YIL variant na.amber.bsc1 — Amber DNA parameters, bsc1 variant na.charmm.c36 — Charmm36 nucleic acid forcefield water.spc — spc 3-site water model water.spc_opls — spc 3-site water model with geometric combining rules for compatability with opls based forcefields water.spce — spce 3-site water model water.tip3p — tip3p 3-site water model water.tip3p-fb — tip3p-fb 3-site water model water.tip3p_charmm — tip3p 3-site water model with additional hydrogen vdw parameters. Used by charmm water.tip4p — tip4p 4-site water model water.tip4p2005 — tip4p-2005 4-site water model water.tip4pd — tip4pd 4-site water model water.tip4pd-1.6 — tip4pd-1.6 water model, for use with aa.amber-disp water.tip4pew — tip4pew 4-site water model water.tip5p — tip5p 5-site water model
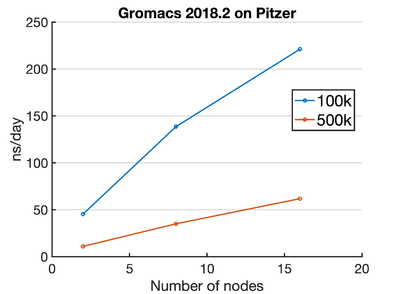
Pitzer
We participated in the Pitzer Early Program, which will become open to the public from Dec 4 2018.
Here is a page on migrating your job from Owens to Pitzer.
Introduction to Pitzer queues and batch limits
Here is a standard Pitzer Gromacs job script:
#PBS -N Ups1-NN
#PBS -l nodes=10:ppn=40
#PBS -l walltime=24:00:00
#PBS -S /bin/bash
#PBS -j oe
#PBS -A PAS1326
trap "cd $PBS_O_WORKDIR; rsync -avh $TMPDIR/ .; exit" TERM
date
module load gromacs
# PBS_O_WORKDIR refers to the directory from which the job was submitted.
cd $PBS_O_WORKDIR
prefix=step5_md
gmx convert-tpr -s ${prefix}-MM.tpr -o ${prefix}-NN.tpr -extend 200000
pbsdcp -p ${prefix}-MM.cpt ${prefix}-NN.tpr $TMPDIR
# Use TMPDIR for best performance.
cd $TMPDIR
mpiexec gmx_mpi mdrun -v -noappend -ntomp 1 -cpi ${prefix}-MM.cpt -deffnm ${prefix}-NN
pbsdcp -p * $PBS_O_WORKDIR/
Things to note:
- The trap command is for copying out files when terminating
- -noappend is required for Gromacs/2018.2 which is default on Pitzer
- -ntomp 1 is A MUST on both Owens and Pitzer, might not be stated on the software page but confirmed by the OSC staff
Submit dependent jobs using this
Benchmarks
!!!If you are getting significantly lower numbers than these, talk to me -Kev!!!
| (ns/day) | Amber16 | Gromacs 2019 | Gromacs 2020 | NAMD 2.13 |
|---|---|---|---|---|
| V100 | ||||
| 90-100k | 184.9 | 58.0 | ||
| 400k | 53.4 | 13.5 [1] | 36.8 [1] | |
| 1M | 15.2 [2] | 3.8 [1] | 10.8 [1] | 6.0 |
| RTX 2080 Ti | ||||
| 90-100k | 137.0 | 98.0 | ||
| 165-170k | 60.0 | 64.0 | ||
| GTX 1080 | ||||
| 90-100k | 43.6 | 48.5 | ||
| 170k | 26.1 | 27.3 | ||
Exxact machines @UC
Computers @R644
[1] 4 x V100 SXM2 16GB
[2] HMR 4fs
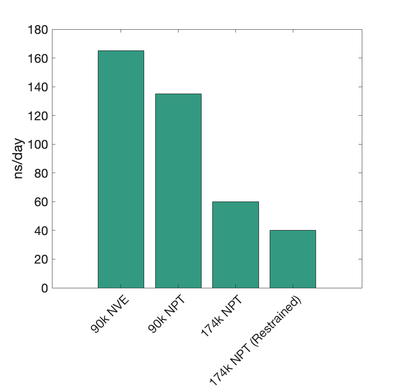
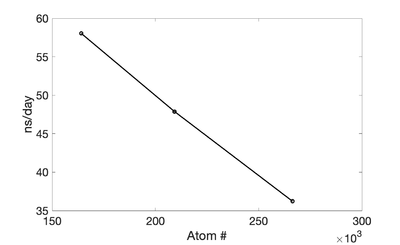
Exxact
(Updated on 3/6/2020 -Kev)
1 GPU (NVIDIA RTX 2080 Ti)